
Optical imaging + computer vision
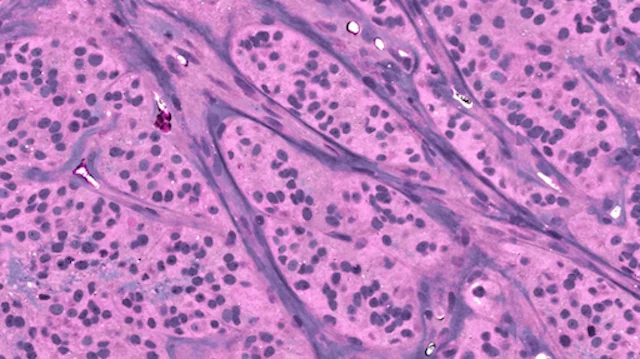
A major focus of the MLiNS lab is to combine stimulated Raman histology (SRH), a rapid label-free, optical imaging method, with deep learning and computer vision techniques to discover the molecular, cellular, and microanatomic features of skull base and malignant brain tumors. We are using SRH in our operating rooms to improve the speed and accuracy of brain tumor diagnosis. Our group has focused on deep learning-based computer vision methods for automated image interpretation, intraoperative diagnosis, and tumor margin delineation. Our work culminated in a multicenter, prospective, clinical trial, which demonstrated that AI interpretation of SRH images was equivalent in diagnostic accuracy to pathologist interpretation of conventional histology. We were able to show, for the first time, that a deep neural network is able to learn recognizable and interpretable histologic image features (e.g. tumor cellularity, nuclear morphology, infiltrative growth pattern, etc) in order to make a diagnosis. Our future work is directed at going beyond human-level interpretation towards identifying molecular/genetic features, single-cell classification, and predicting patient prognosis.
Featured Publications
CVPR | 2023
Hierarchical Discriminative Learning Improves Visual Representations of Biomedical MicroscopyNATURE MEDICINE | 2023
AI-based molecular classification of diffuse gliomas using rapid, label-free optical imagingNEURIPS DATASETS & BENCHMARKS | 2022
OpenSRH: optimizing brain tumor surgery using intraoperative stimulated Raman histologyNEUROSURGERY | 2022
Rapid automated analysis of skull base tumor specimens using intraoperative optical imaging and artificial intelligenceNEURO-ONCOLOGY | 2021
Rapid, label-free detection of diffuse glioma recurrence using intraoperative stimulated Raman histology and deep neural networksNATURE MEDICINE | 2020
Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networksCANCER RESEARCH | 2018
Rapid Intraoperative Diagnosis of Pediatric Brain Tumors Using Stimulated Raman HistologyNATURE BIOMEDICAL ENGINEERING | 2017
Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopyJOURNAL OF NEUROSURGERY | 2016
Improving the accuracy of brain tumor surgery via Raman-based technology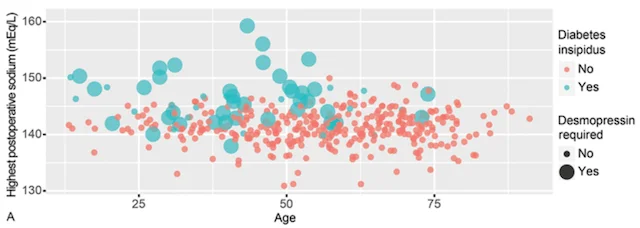
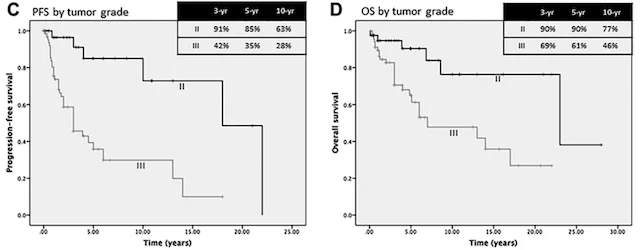

Predicting patient outcomes
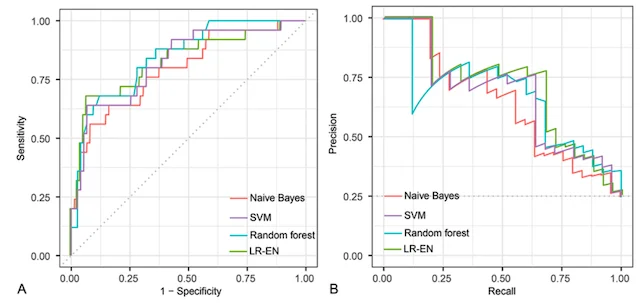
Machine learning has the potential to revolutionize the way we predict patient outcomes. Electronic medical records (EMR) have streamlined data collection and made it easier than ever to use ML algorithms to assist in patient care. Our work has focused on predicting (1) how well patients will recover after surgery and (2) what will ultimately affect their long-term outcome and survival. We use ML algorithms that allow for transparency and interpretability, both of which are essential to trust the recommendations of ML decision-support tools in healthcare. By identifying the specific set of symptoms, radiographic findings, laboratory values, etc. that our models use for prediction, physicians can use ML recommendations in the appropriate clinical context for personalized treatment decisions.
Featured Publications
JOURNAL OF NEUROSURGERY | 2018
A machine learning approach to predict early outcomes after pituitary adenoma surgeryCRITICAL CARE MEDICINE | 2018
Clinical Factors Associated With ICU-Specific Care Following Supratentoral Brain Tumor Resection and Validation of a Risk Prediction ScoreJOURNAL OF NEUROSURGERY | 2016
Supratentorial hemispheric ependymomas: an analysis of 109 adults for survival and prognostic factors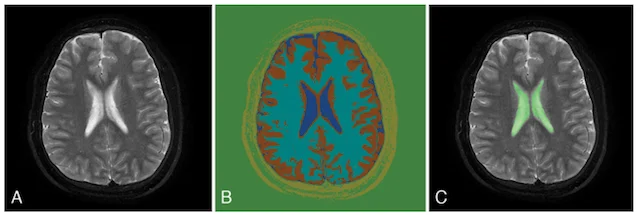


Automated brain segmentation
Computed tomography (CT) and magnetic resonance imaging (MRI) are an indispensable part of neurosurgery. Currently, interpretation of CT and MRI scans requires a skilled neuroradiologist and is mainly qualitative, meaning the radiologist provides a written narrative of their findings with little or no quantitative measurements. These measurements are time- and labor-intensive, making it infeasible to have neuroradiologists perform them in the vast majority of scans. Therefore, the MLiNS lab has set out to automate the process of measuring specific features of MR and CT scans that can affect treatment decisions and patient outcomes. For example, the volume of the cerebral ventricles is an essential part of the diagnosis and long-term management of hydrocephalus. The problem is that (1) cerebral ventricle volumes can vary widely between patients and (2) detecting subtle changes in ventricle volume can be challenging, even by trained neuroradiologists and neurosurgeons. We have developed a method for automated segmentation and volumetric assessment of cerebral ventricles in both normal controls and hydrocephalus patients to assist in treatment decisions. We have applied similar methods to measure the hematoma burden in subarachnoid hemorrhage patients and morphologic assessment of the posterior fossa in Chiari malformations.
Featured Publications
PITUITARY | 2022
Generating novel pituitary datasets from open-source imaging data and deep volumentric segmentationJOURNAL OF NEUROSURGERY PEDIATRICS | 2020
Normal cerebral ventricular volume growth in childhoodJOURNAL OF NEUROSURGERY | 2020
Volumetric quantification of aneurysmal subarachnoid hemorrhage independently predicts hydrocephalus and seizuresJOURNAL OF NEUROSURGERY PEDIATRICS | 2017
Morphometric and volumetric comparison of 102 children with symptomatic and asymptomatic Chiari malformation Type IJOURNAL OF NEUROSURGERY PEDIATRICS | 2017
Comparison of posterior fossa volumes and clinical outcomes after decompression of Chiari malformation Type I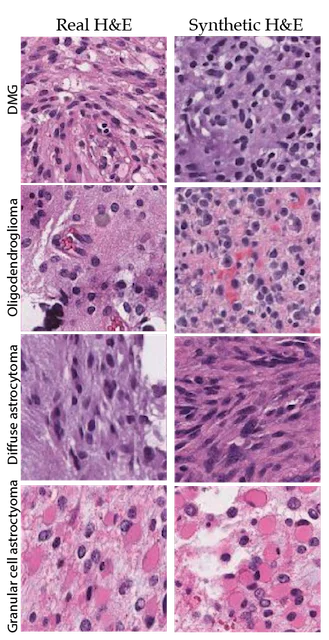
Augmenting intraoperative pathology
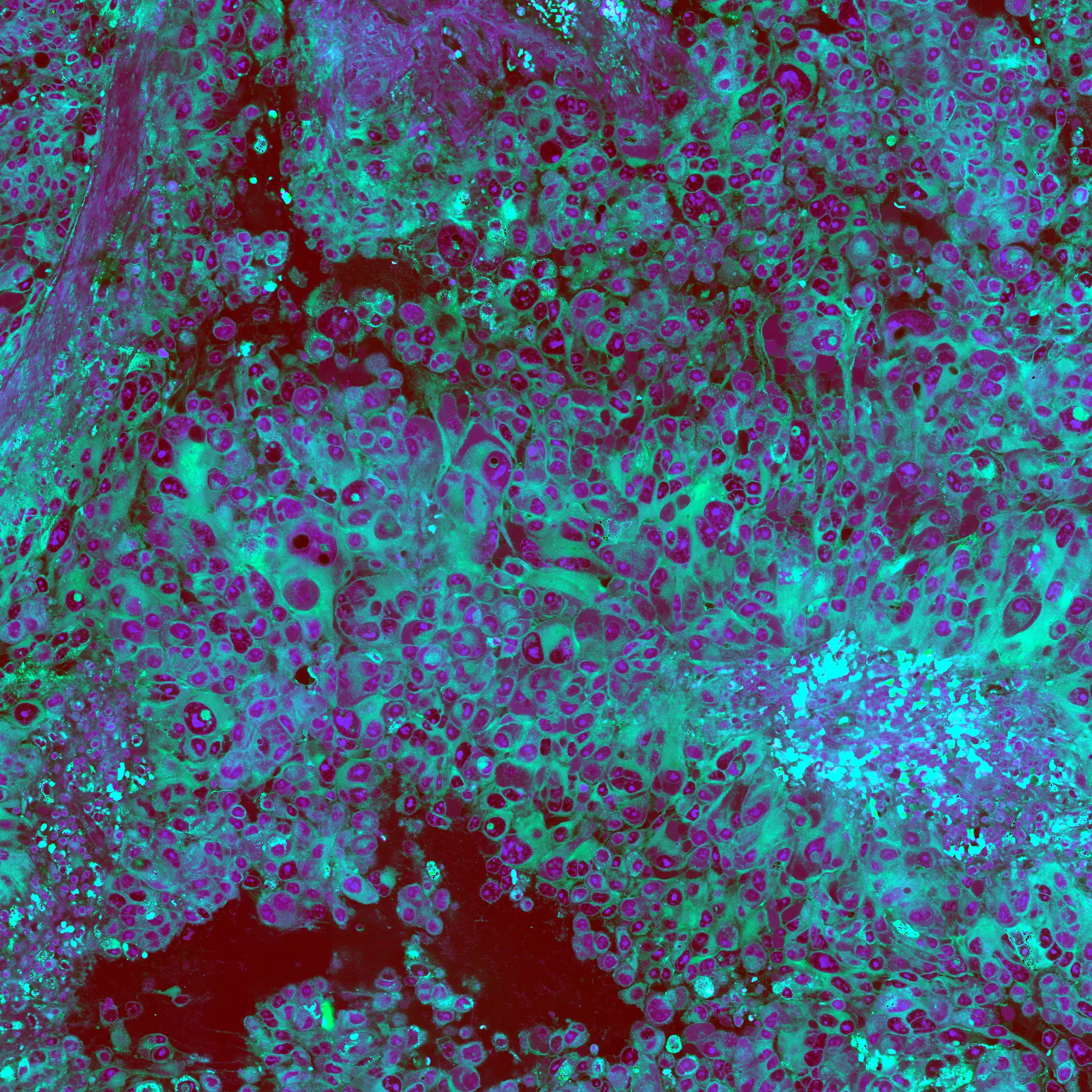
The use of AI in digital pathology has grown significantly over the previous decade. The majority of research in this area has focused on the automated interpretation of formalin-fixed, paraffin-embedded permanent specimens. However, our lab has focused on applying AI to frozen sections performed during surgery, which is a much more challenging computer vision task. Accurate intraoperative diagnosis is essential for providing safe and effective care for brain tumor patients. Surgical goals are divergent depending on intraoperative diagnosis, which necessitates a pathology workflow that is timely and accurate. Conventional intraoperative pathology requires freezing and sectioning, which introduces artifacts into the specimen and can make interpretation challenging and decrease accuracy. Moreover, each fresh intraoperative specimen must be individually reviewed by a neuropathologist, which can be time-consuming. Our lab aims to provide decision support and computer-augmented diagnostic tools to improve the speed and accuracy intraoperative diagnosis. Specifically, we have trained deep neural networks to classify the most common brain tumor types using frozen sections alone. Because the availability of frozen whose-slide images is limited, we use synthetic images for data augmentation to improve classification performance. We train a neural network model, called a generative adversarial network, to generate 'fake' images that can be used to train a classifier network to improve diagnostic accuracy. We have worked closely on this project with our collaborators at Synthetaic, a start-up AI company that uses synthetic data to improve AI for real-world applications.
The images on the left are real images sampled from the The Cancer Genome Atlas. The images on the right are synthetic, or fake, images created using generative adversarial networks. We use the fake images to improve brain tumor diagnosis.
Featured Publications
CNS ONCOLOGY | 2020
Automated histologic diagnosis of CNS tumors with machine learning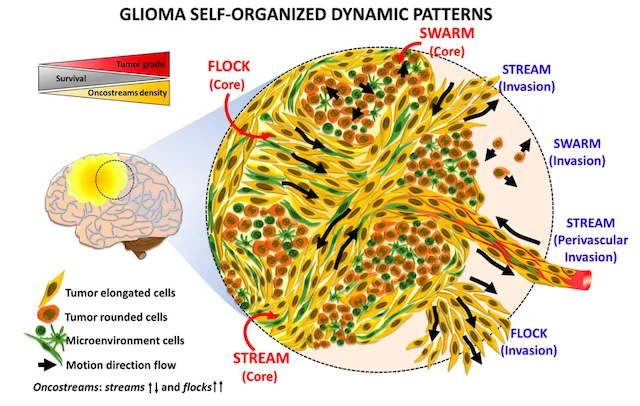
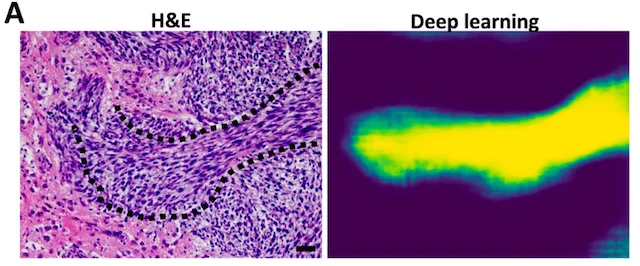
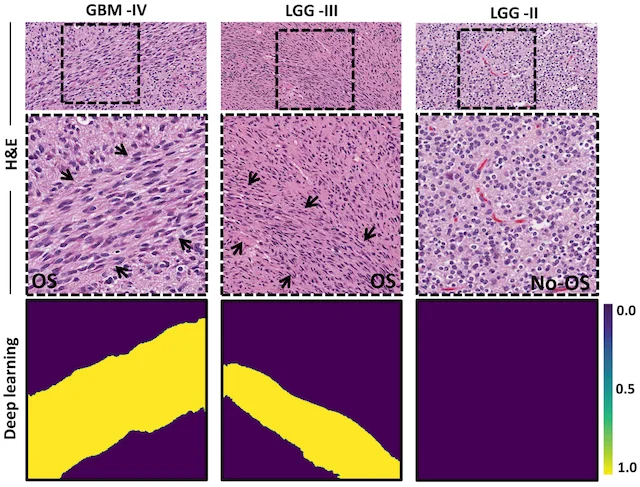
Finding oncostreams in gliomas
Our collaborators, Drs. Pedro Lowenstein and Maria Castro, have discovered that within gliomas are self-organizing structures, called oncostreams, which determine the spatial heterogeneity and malignant behavior of these deadly tumors. The MLiNS lab was able to assist in this discovery by developing deep neural networks that can detect and quantify oncostreams in whole-slide images from glioma mouse models and patient's tumors. Using the latest computer vision methods for biomedical image segmentation, we were able to demonstrate that a higher density of oncostreams, detected using our trained model, correlates with tumor grade and decreased survival. These results were confirmed using whole-slide images from The Cancer Genome Atlas (TCGA).
Featured Publications
NATURE COMMUNICATIONS | 2022
Spatiotemporal analysis of glioma heterogeneity reveals COL1A1 as an actionable target to disrupt tumor progressionFRONTIERS IN ONCOLOGY | 2021
Uncovering Spatiotemporal Heterogeneity of High-Grade Gliomas: From Disease Biology to Therapeutic Implications