CVPR WORKSHOP ON COMPUTER VISION FOR MICROSCOPY IMAGE ANALYSIS | 2024
Super-resolution of biomedical volumes with 2D supervision
Cheng Jiang1*, Alexander Gedeon1*, Yiwei Lyu1, Eric Landgraf1, Yufeng Zhang1, Xinhai Hou1, Akhil Kondepudi1, Asadur Chowdury1, Honglak Lee1, and Todd C. Hollon1
1University of Michigan *Equal contribution
MSDSR super-resolves biomedical volumes without training with high resolution 3D volumes by leveraging spatial dimension equivalence in biological specimens.
Abstract
Volumetric biomedical microscopy has the potential to increase the diagnostic information extracted from clinical tissue specimens and improve the diagnostic accuracy of both human pathologists and computational pathology models. Unfortunately, barriers to integrating 3-dimensional (3D) volumetric microscopy into clinical medicine include long imaging times, poor depth/z-axis resolution, and an insufficient amount of high-quality volumetric data. Leveraging the abundance of high-resolution 2D microscopy data, we introduce masked slice diffusion for super-resolution (MSDSR), which exploits the inherent equivalence in the data-generating distribution across all spatial dimensions of biological specimens. This intrinsic characteristic allows for super-resolution models trained on high-resolution images from one plane (e.g., XY) to effectively generalize to others (XZ, YZ), overcoming the traditional dependency on orientation. We focus on the application of MSDSR to stimulated Raman histology (SRH), an optical imaging modality for biological specimen analysis and intraoperative diagnosis, characterized by its rapid acquisition of high-resolution 2D images but slow and costly optical z-sectioning. To evaluate MSDSR's efficacy, we introduce a new performance metric, SliceFID, and demonstrate MSDSR's superior performance over baseline models through extensive evaluations. Our findings reveal that MSDSR not only significantly enhances the quality and resolution of 3D volumetric data, but also addresses major obstacles hindering the broader application of 3D volumetric microscopy in clinical diagnostics and biomedical research.
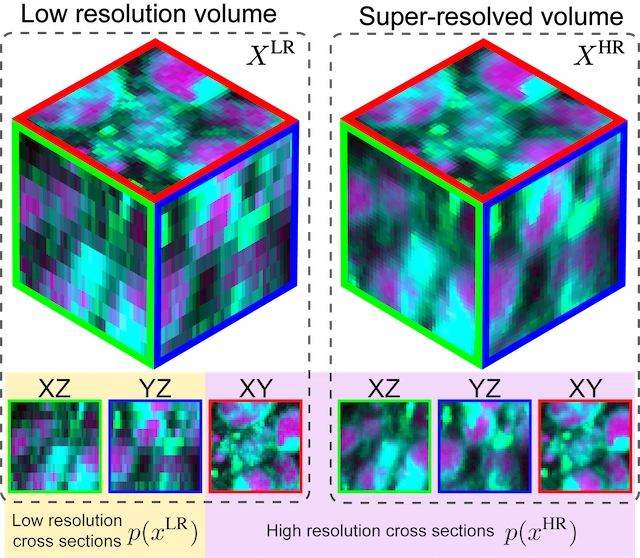
Super-resolution of biomedical volumes with 2D supervision. Volumetric microscopy images have a data distribution agnostic to tissue orientation and spatial dimension. This intrinsic characteristic allows for super-resolution models trained on high-resolution images from one plane (e.g., XY) to effectively generalize to others (XZ, YZ), overcoming the traditional dependency on orientation. Here, we present a method for leveraging this intrinsic characteristic by super-resolving low-resolution volumes using a conditional diffusion model trained on 2D high-resolution images.
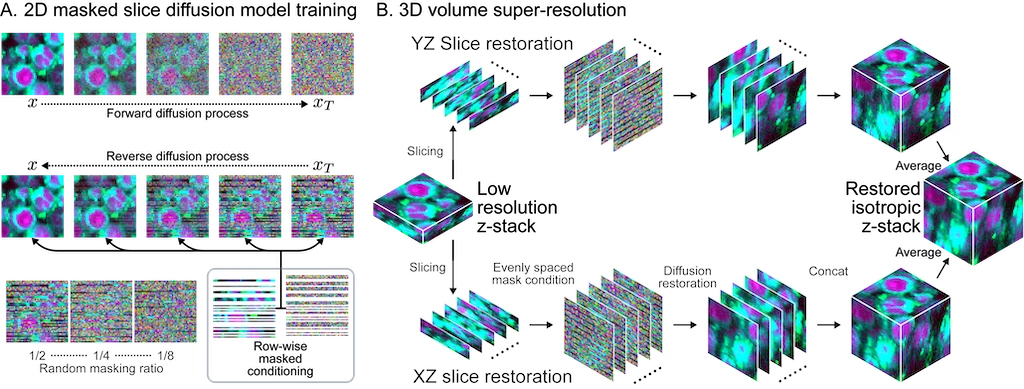
MSDSR overview
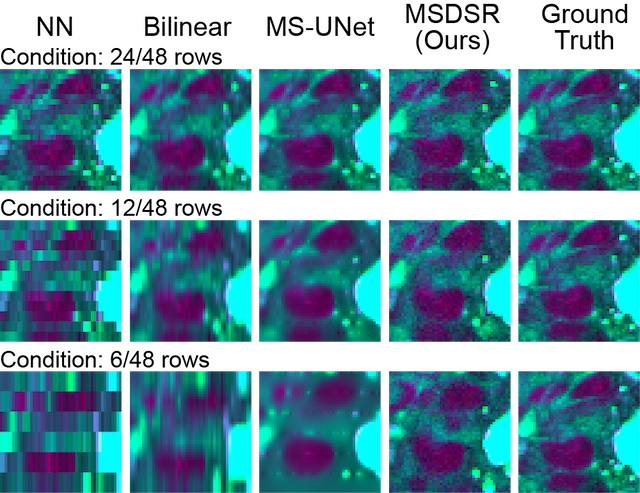
2D evaluation results
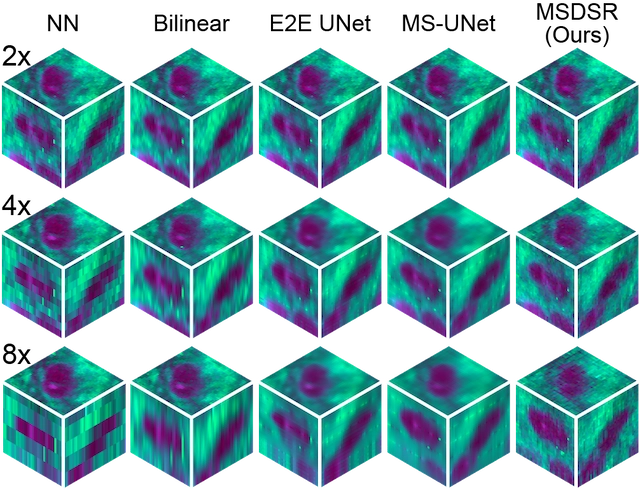
3D evaluation results
Bibtex
@inproceedings{jiang2024super,
title={Super-resolution of biomedical volumes with 2D supervision},
author={Jiang, Cheng and Gedeon, Alexander and Lyu, Yiwei and Landgraf, Eric and Zhang, Yufeng and Hou, Xinhai and Kondepudi, Akhil and Chowdury, Asadur and Lee, Honglak and Hollon, Todd},
booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops},
year={2024}
}
