Selected as a highlight at CVPR 2023
Hierarchical Discriminative (HiDisc) Learning Improves Visual Representations of Biomedical Microscopy
Cheng Jiang1*, Xinhai Hou1*, Akhil Kondepudi1, Asadur Chowdury1, Christian W. Freudiger2, Daniel A. Orringer3, Honglak Lee1,4, and Todd C. Hollon1
1University of Michigan 2Invenio Imaging 3New York University 4LG AI Research *Equal Contribution
Abstract
Learning high-quality, self-supervised, visual representations is essential to advance the role of computer vision in biomedical microscopy and clinical medicine. Previous work has focused on self-supervised representation learning (SSL) methods developed for instance discrimination and applied them directly to image patches, or fields-of-view, sampled from gigapixel whole-slide images (WSIs) used for cancer diagnosis. However, this strategy is limited because it (1) assumes patches from the same patient are independent, (2) neglects the patient-slide-patch hierarchy of clinical biomedical microscopy, and (3) requires strong data augmentations that can degrade downstream performance. Importantly, sampled patches from WSIs of a patient's tumor are a diverse set of image examples that capture the same underlying cancer diagnosis. This motivated HiDisc, a method that leverages the inherent patient-slide-patch hierarchy of clinical biomedical microscopy to define a hierarchical discriminative learning task that implicitly learns features of the underlying diagnosis. HiDisc uses a self-supervised contrastive learning framework in which positive patch pairs are defined based on a common ancestry in the data hierarchy, and a unified patch, slide, and patient discriminative learning objective is used for visual SSL. We benchmark HiDisc visual representations on two vision tasks using two biomedical microscopy datasets, and demonstrate that (1) HiDisc pretraining outperforms current state-of-the-art self-supervised pretraining methods for cancer diagnosis and genetic mutation prediction, and (2) HiDisc learns high-quality visual representations using natural patch diversity without strong data augmentations.
Motivation
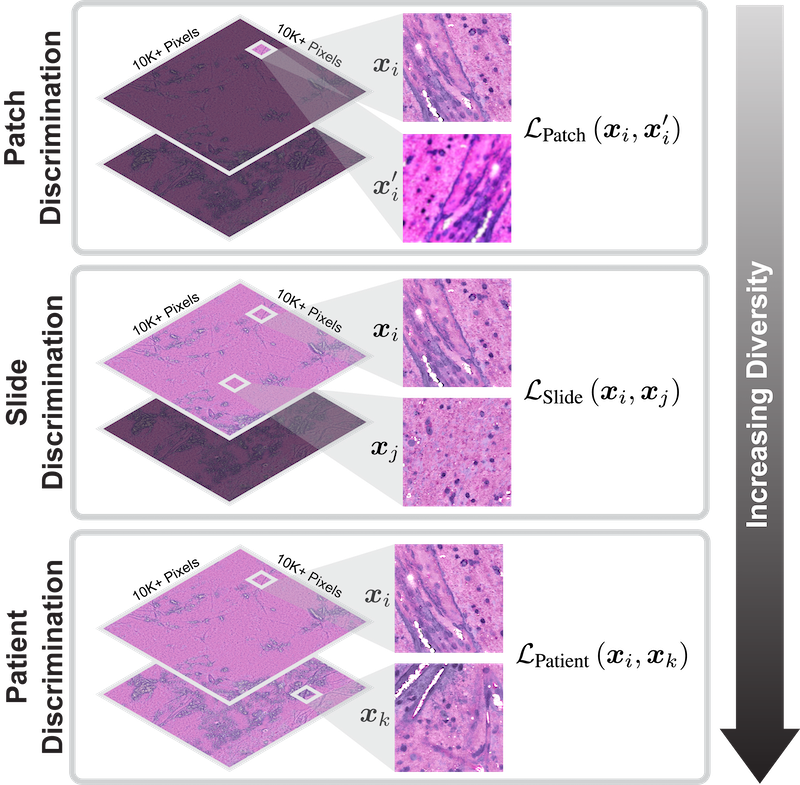
Clinical biomedical microscopy has a hierarchical patch-slide-patient data structure. HiDisc combines patch, slide, and patient discrimination into a unified self-supervised learning task.
Algorithm pseudocode

Hierarchical discriminative learning overview
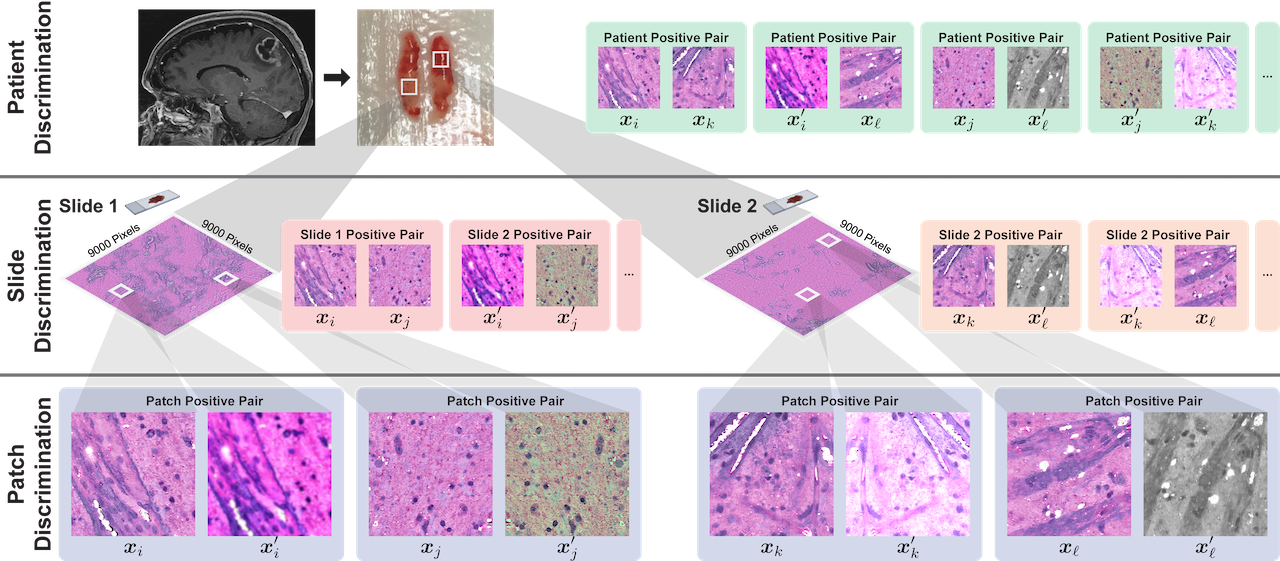
Motivated by the patient-slide-patch data hierarchy of clinical biomedical microscopy, HiDisc defines a patient, slide, and patch discriminative learning objective to improve visual representations. Because WSI and microscopy data are inherently hierarchical, defining a unified hierarchical loss function does not require additional annotations or supervision. Positive patch pairs are defined based on a common ancestry in the data hierarchy. A major advantage of HiDisc is the ability to define positive pairs without the need to sample from or learn a set of strong image augmentations, such as random erasing, shears, color inversion, etc. Because each field-of-view in a WSI is a different view of a patient's underlying cancer diagnosis, HiDisc implicitly learns image features that predict that diagnosis.
Visualization of learned SRH representations using SimCLR and HiDisc
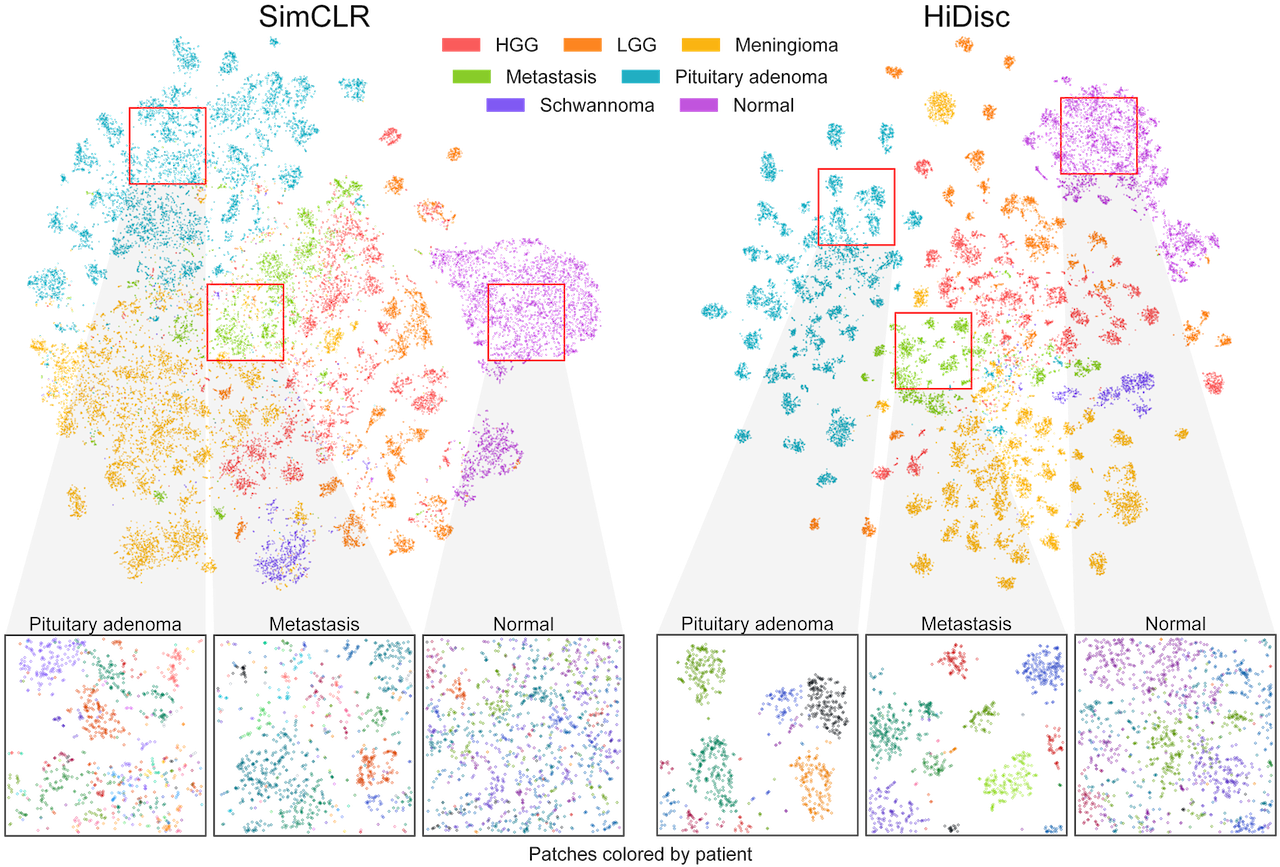
Top. Randomly sampled patch representations are visualized after SimCLR versus HiDisc pretraining using tSNE. Representations are colored based on brain tumor diagnosis. HiDisc qualitatively achieves higher quality feature learning and class separation compared to SimCLR. Expectedly, HiDisc shows within- diagnosis clustering that corresponds to patient discrimination. Bottom. Magnified cropped regions of the above visualizations show subclusters that correspond to individual patients. Patch representations in magnified crops are colored according to patient membership. We see patient discrimination within the different tumor diagnoses. Importantly, we do not see patient discrimination within normal brain tissue because there are minimal-to-no differentiating microscopic features between patients. This demonstrates that in the absence of discriminative features at the slide- or patient-level, HiDisc can achieve good feature learning using patch discrimination without overfitting the other discrimination tasks.
Bibtex
@inproceedings{jiang2023hierarchical,
title={Hierarchical discriminative learning improves visual representations of biomedical microscopy},
author={Jiang, Cheng and Hou, Xinhai and Kondepudi, Akhil and Chowdury, Asadur Zaman and Freudiger, Christian and Orringer, Daniel A and Lee, Honglak and Hollon, Todd},
booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition},
year={2023}
}
