Protocol to Annotate and Train Deep Learning Models to Automate Single Cell Detection and Segmentation on Stimulated Raman Histology (SRH)
Abhishek Bhattacharya*1,2, Eric Landgraf*1, Cheng Jiang1, Asadur Chowdury1, Akhil Kondepudi1, Lin Wang1, Edward S. Harake1, Xinhai Hou1, Lisa Walsh1, and Todd C. Hollon1
1University of Michigan 2New York University *Equal Contribution
Summary
Stimulated Raman Histology (SRH) is a label-free optical imaging technique that can discern molecular components such as lipids and proteins at a subcellular spatial resolution without histologic staining. This method is used intra-operatively for rapid imaging of human tissue biopsies during neurosurgical cases. To enable single cell spatial analysis on SRH, we present a pipeline for labeling cells and training AI models for automated cell segmentation using ELUCIDATE, a web-based SRH cell annotation tool, and DetectSRH Python library.
Highlights
- Step-by-step workflow from manual labeling to automated cell detection and analysis
- A web-based platform for collaborative cell annotation on Stimulated Raman Histology (SRH)
- Expert-annotated single cell segmentation dataset from multiple brain tumor types
- A Python library for training deep learning instance segmentation models on SRH and associated open source model weights
ELUCIDATE
Annotation interface
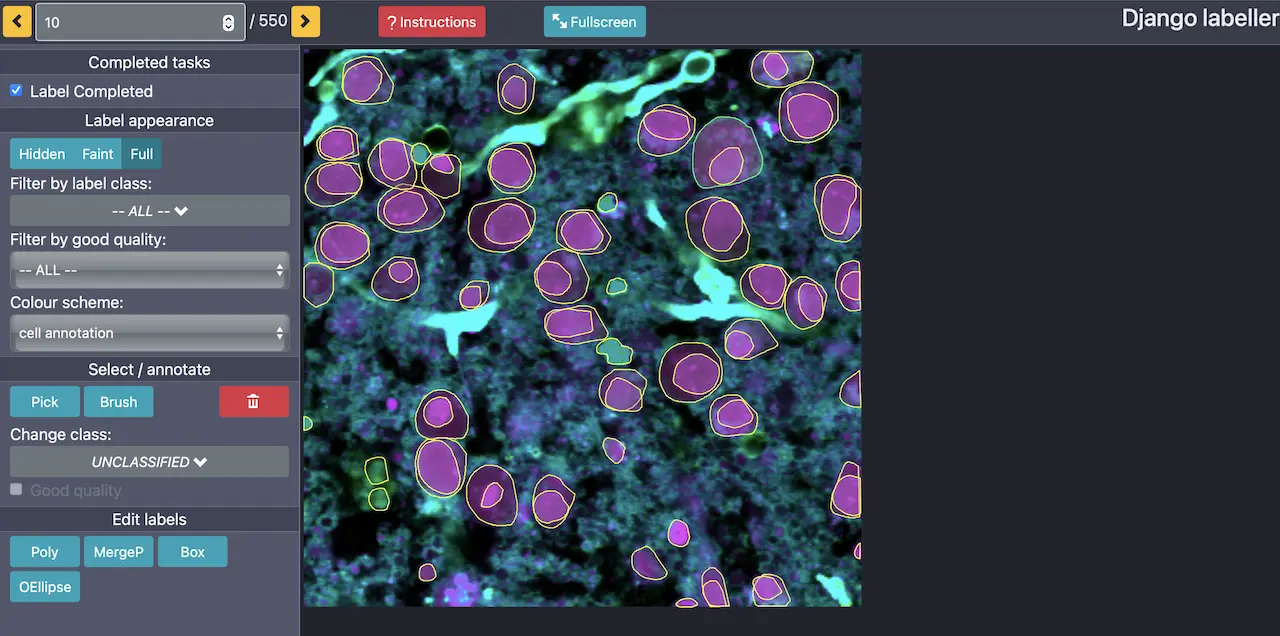
The annotation viewer allows you to view and create polygonal segmentations (using the panel on the left) with assigned labels over any part of the image. The x, y coordinates with associated labels will be saved automatically and available for export. You can pan around the image by right-clicking and moving the blank space on the outside in the desired direction. Detailed instructions for how to use the other features in the annotation viewer can be seen by clicking the red instructions button at the top of the interface.
Building SRH Annotation Database Using Model Assistance
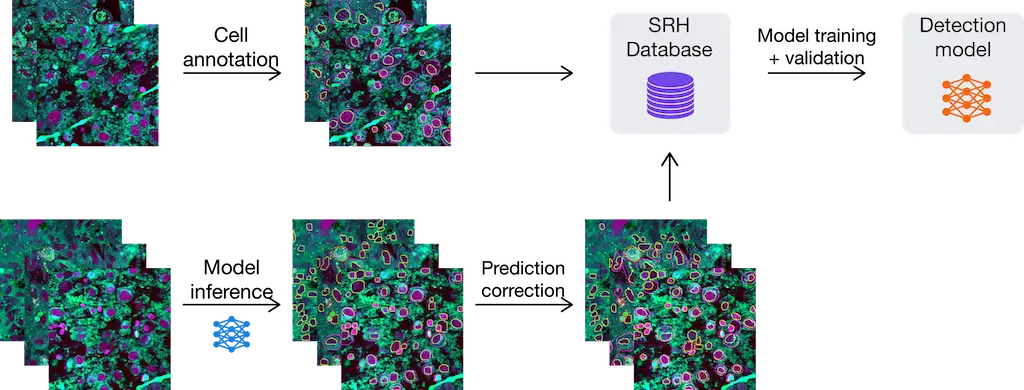
The diagram illustrates the manual (above) and model-assisted methods (below) workflow for building a large SRH database of cell annotations. The workflow below allows for correcting model predictions directly on ELUCIDATE before feeding it in for model training and validation, which can be repeated as needed.
SRH550
Annotation statistics

Left, number of annotations per cell type or cell structure label. Right, Number of annotations per tumor type. Abbreviations: HGG, high grade glioma; LGG, low grade glioma; Mening, meningioma; Mets, metastasis; Pit, pituitary adenoma; Schwan, schwannoma; Normal, normal brain.
DetectSRH
Nuclei instance segmentation metrics
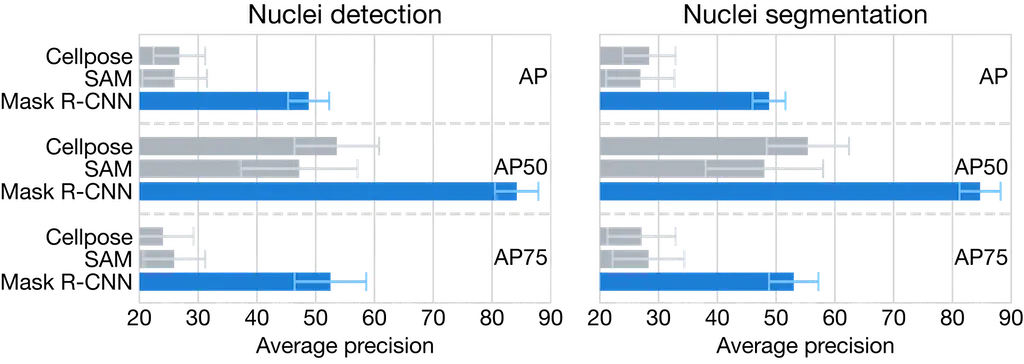
Nuclei instance segmentation AP50 for all methods across all tumor types radar plot. The shaded area represents the standard deviation across 5 fold cross-validation. Abbreviations: HGG, high grade glioma; LGG, low grade glioma; Mening, meningioma; Mets, metastasis; Pit, pituitary adenoma; Schwan, schwannoma; Normal, normal brain.
Nuclei instance segmentation visualization
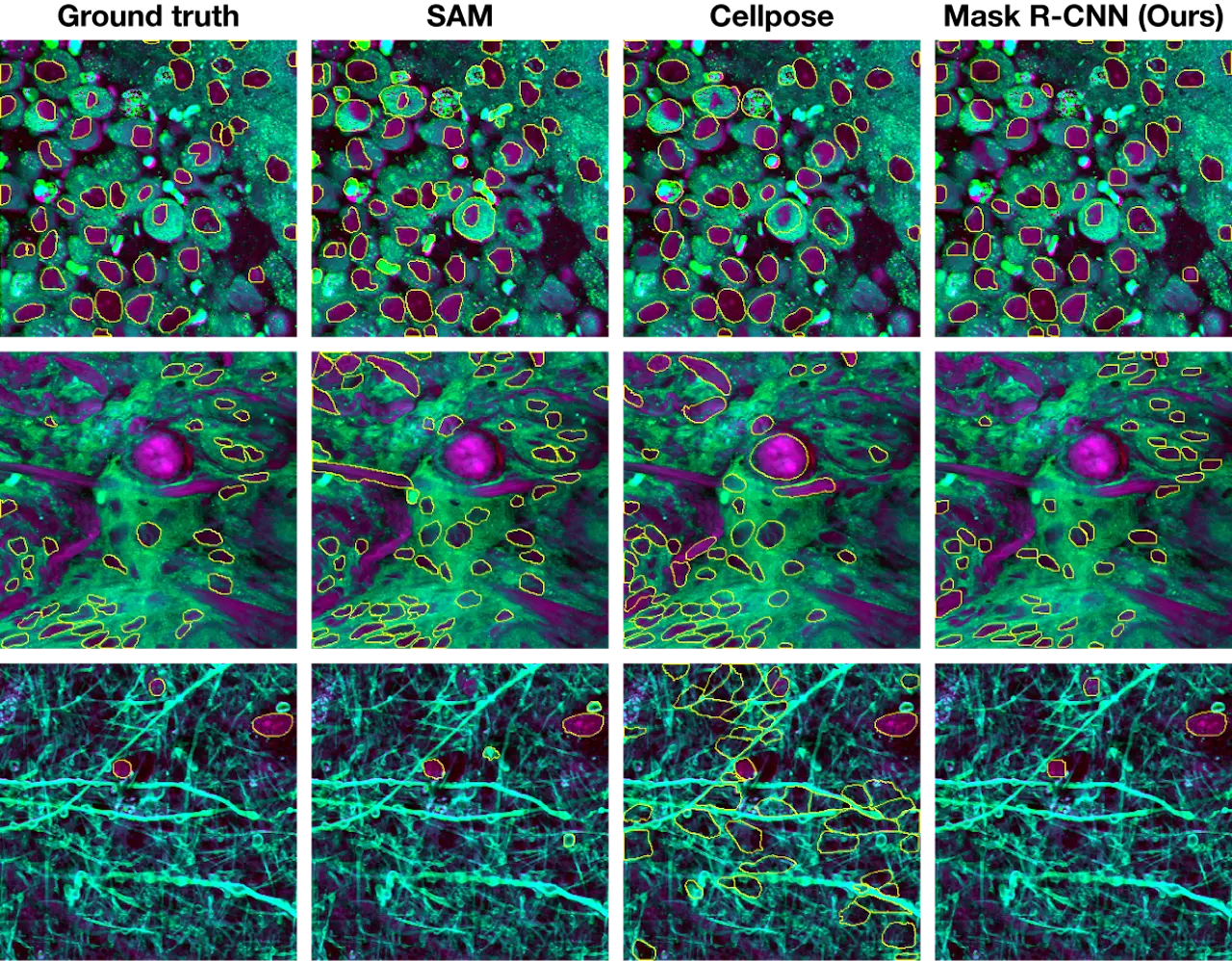
Comparison of nuclei segmentation results from SAM, Cellpose, and Mask R-CNN models against ground truth annotations.
Nuclei instance segmentation metriccs per tumor type
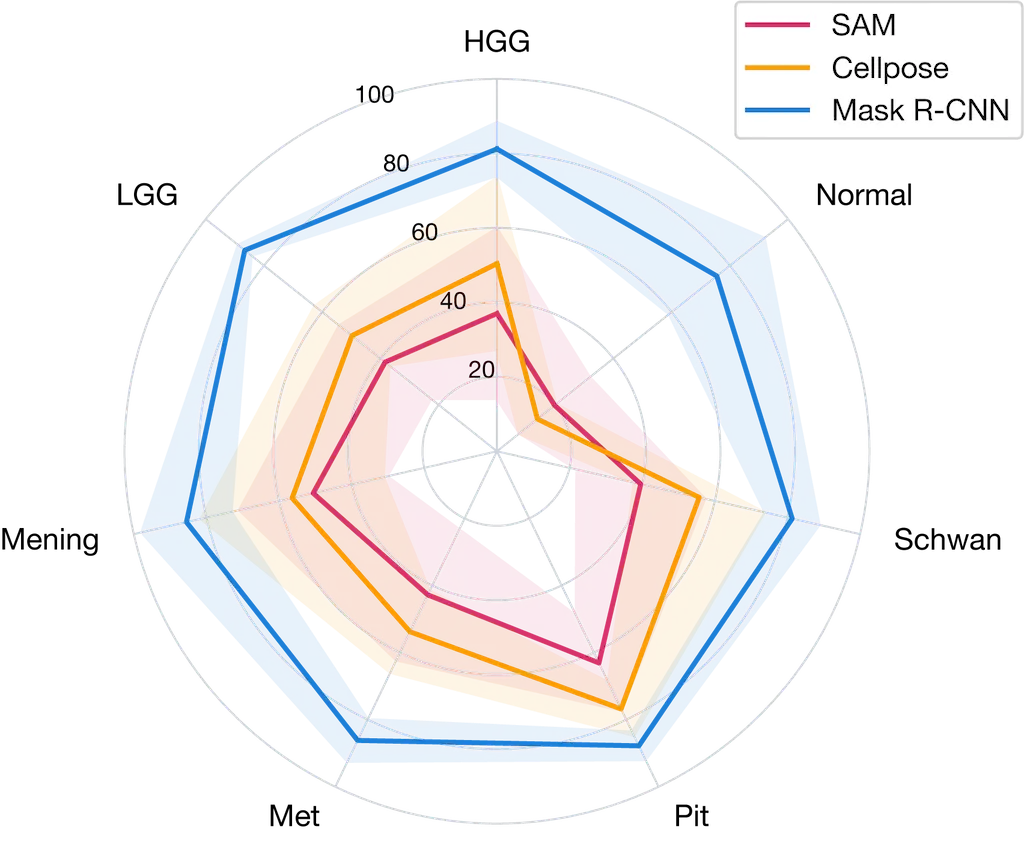
Nuclei instance segmentation AP50 for all methods across all tumor types radar plot. The shaded area represents the standard deviation across 5 fold cross-validation. Abbreviations: HGG, high grade glioma; LGG, low grade glioma; Mening, meningioma; Mets, metastasis; Pit, pituitary adenoma; Schwan, schwannoma; Normal, normal brain.
Cell instance segmentation metrics
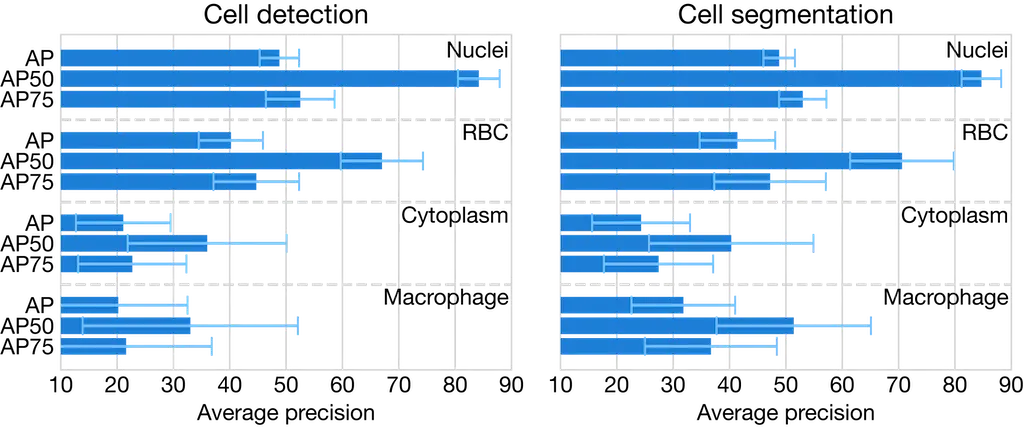
Mask R-CNN instance segmentation performance metrics. Detection and segmentation average precision (AP) scores across cell types with standard deviations from 5-fold cross-validation. AP scores shown for nuclei, red blood cells (RBC), cytoplasm, and macrophages.
Cell instance segmentation visualization
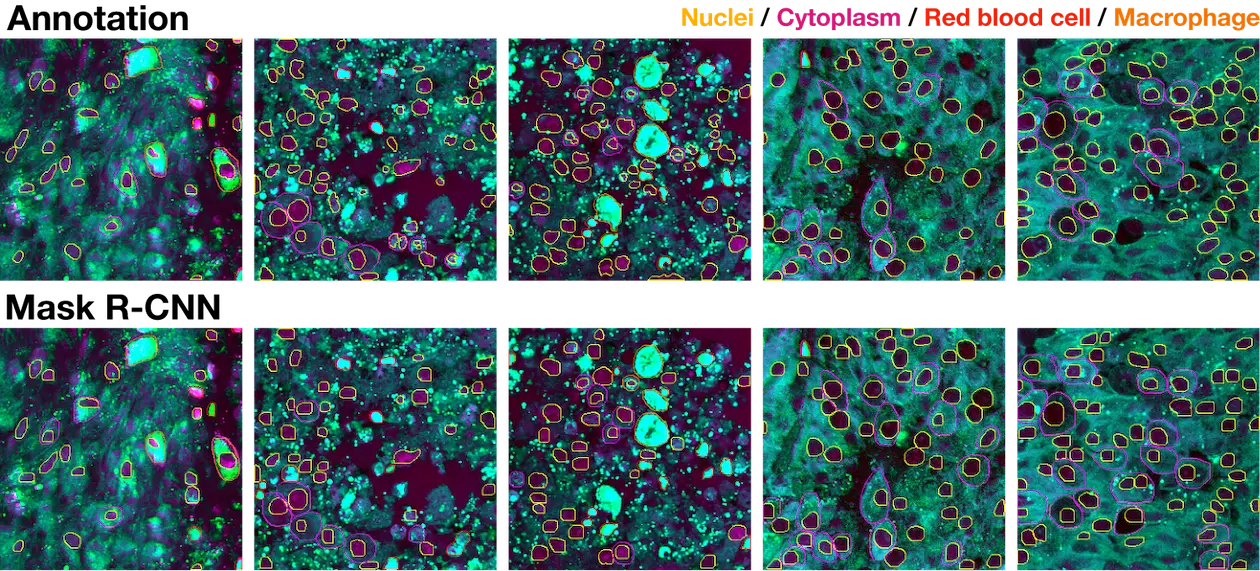
Mask R-CNN segmentation examples of nuclei, cytoplasm, red blood cells, and macrophages with corresponding ground truth annotations.
Bibtex
Coming soon!
